Maximum Likelihood Estimation in R
.jpg)
Often, you’ll have some level of intuition—or perhaps concrete evidence—to suggest that a set of observations has been generated by a particular statistical distribution. Similar phenomena to the one you are modelling may have been shown to be explained well by a certain distribution. The setup of the situation or problem you are investigating may naturally suggest a family of distributions to try. Or maybe you just want to have a bit of fun by fitting your data to some obscure model just to see what happens (if you are challenged on this, tell people you’re doing Exploratory Data Analysis and that you don’t like to be disturbed when you’re in your zone).
Now, there are many ways of estimating the parameters of your chosen model from the data you have. The simplest of these is the method of moments—an effective tool, but one not without its disadvantages (notably, these estimates are often biased).
Another method you may want to consider is Maximum Likelihood Estimation (MLE), which tends to produce better (ie more unbiased) estimates for model parameters. It’s a little more technical, but nothing that we can’t handle. Let’s see how it works.
What is likelihood?
The likelihood—more precisely, the likelihood function—is a function that represents how likely it is to obtain a certain set of observations from a given model. We’re considering the set of observations as fixed—they’ve happened, they’re in the past—and now we’re considering under which set of model parameters we would be most likely to observe them.
A simple coin-flipping example
Consider an example. Let’s say we flipped a coin 100 times and observed 52 heads and 48 tails. We want to come up with a model that will predict the number of heads we’ll get if we kept flipping another 100 times.
Formalising the problem a bit, let’s denote the random variable X as being the number of heads obtained from 100 coin flips. Given that:
there are only two possible outcomes (heads and tails),
there’s a fixed number of “trials” (100 coin flips), and that
there’s a fixed probability of “success” (ie getting a heads),
we might reasonably suggest that the situation could be modelled using a binomial distribution.
We can use R to set up the problem as follows:
# I don't know about you but I'm feeling
set.seed(22)
# Generate an outcome, ie number of heads obtained, assuming a fair coin was
# used for the 100 flips
heads <- rbinom(1,100,0.5)
heads
# 52(For the purposes of generating the data, we’ve used a 50/50 chance of getting a heads/tails, although we are going to pretend that we don’t know this for the time being. For almost all real world problems we don’t have access to this kind of information on the processes that generate the data we’re looking at—which is entirely why we are motivated to estimate these parameters!)
Under our formulation of the heads/tails process as a binomial one, we are supposing that there is a probability p of obtaining a heads for each coin flip. Extending this, the probability of obtaining 52 heads after 100 flips is given by:
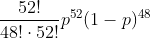
This probability is our likelihood function—it allows us to calculate the probability, ie how likely it is, of that our set of data being observed given a probability of heads p. You may be able to guess the next step, given the name of this technique—we must find the value of p that maximises this likelihood function.
We can easily calculate this probability in two different ways in R:
# To illustrate, let's find the likelihood of obtaining these results if p was
# 0.6—that is, if our coin was biased in such a way to show heads 60%
# of the time.
biased_prob <- 0.6
# Explicit calculation
choose(100,52)*(biased_prob**52)*(1-biased_prob)**48
# 0.0214877567069514# Using R's dbinom function (density function for a given binomial distribution)
dbinom(heads,100,biased_prob)
# 0.0214877567069514Back to our problem—we want to know the value of p that our data implies. For simple situations like the one under consideration, it’s possible to differentiate the likelihood function with respect to the parameter being estimated and equate the resulting expression to zero in order to solve for the MLE estimate of p. However, for more complicated (and realistic) processes, you will probably have to resort to doing it numerically.
Luckily, this is a breeze with R as well! Our approach will be as follows:
Define a function that will calculate the likelihood function for a given value of p; then
Search for the value of p that results in the highest likelihood.
Starting with the first step:
likelihood <- function(p){
dbinom(heads, 100, p)
}
# Test that our function gives the same result as in our earlier example
likelihood(biased_prob)
# 0.0214877567069513And now considering the second step. There are many different ways of optimising (ie maximising or minimising) functions in R—the one we’ll consider here makes use of the nlm function, which stands for non-linear minimisation. If you give nlm a function and indicate which parameter you want it to vary, it will follow an algorithm and work iteratively until it finds the value of that parameter which minimises the function’s value.
You may be concerned that I’ve introduced a tool to minimise a function’s value when we really are looking to maximise—this is maximum likelihood estimation, after all! Fortunately, maximising a function is equivalent to minimising the function multiplied by minus one. If we create a new function that simply produces the likelihood multiplied by minus one, then the parameter that minimises the value of this new function will be exactly the same as the parameter that maximises our original likelihood.
As such, a small adjustment to our function from before is in order:
negative_likelihood <- function(p){
dbinom(heads, 100, p)*-1
}
# Test that our function is behaving as expected
negative_likelihood(biased_prob)
# -0.0214877567069513Excellent—we’re now ready to find our MLE value for p.
nlm(negative_likelihood,0.5,stepmax=0.5)
# $minimum
# -0.07965256
# $estimate
# 0.5199995
# $gradient
# -2.775558e-11
# $code
# 1
# $iterations
# 4The nlm function has returned some information about its quest to find the MLE estimate of p.
$minimum denotes the minimum value of the negative likelihood that was found—so the maximum likelihood is just this value multiplied by minus one, ie 0.07965…;
$estimate is our MLE estimate of p;
$gradient is the gradient of the likelihood function in the vicinity of our estimate of p—we would expect this to be very close to zero for a successful estimate;
$code explains to use why the minimisation algorithm was terminated—a value of 1 indicates that the minimisation is likely to have been successful; and
$iterations tells us the number of iterations that nlm had to go through to obtain this optimal value of the parameter.
This information is all nice to know—but what we really care about is that it’s telling us that our MLE estimate of p is 0.52. We can intuitively tell that this is correct—what coin would be more likely to give us 52 heads out of 100 flips than one that lands on heads 52% of the time?
Conclusion
In this rather trivial example we’ve looked at today, it may seems like we’ve put ourselves through a lot of hassle to arrive at a fairly obvious conclusion. But consider a problem where you have a more complicated distribution and multiple parameters to optimise—the problem of maximum likelihood estimation becomes exponentially more difficult—fortunately, the process that we’ve explored today scales up well to these more complicated problems.
Ultimately, you better have a good grasp of MLE estimation if you want to build robust models—and in my estimation, you’ve just taken another step towards maximising your chances of success—or would you prefer to think of it as minimising your probability of failure?
More info and credits
Andrew Hetherington is an actuary and data enthusiast working in London, UK. All views are Andrew’s own and not those of his employer.
Coin photo by Claudia Schwarz on Unsplash.